Zhongyi (James) Guo 过仲懿🌈
About Me | Education | Publications | Experience | Technical Summary | Personal Life ↗

Get In Touch
outreach at gzy1s dot me
M+P-AMA-DG1J (text me to set up a call)
Skill Set
🤖 Machine Learning
👨🏻💻 Data Science
📊 Statistical Modeling
📝 Grant Proposal Writing
🖥️ High Performance Computing
Certifications
SAS
Coursera Badges & Courses
DeepLearning.AI
IBM AI Engineering
Stanford
Languages
- English
- Mandarin
Log
11/20/2024: Updated research experience. I will update project experience and work experience shortly.
11/12/2024: Minor updates applied to the certification section.
08/12/2024: I redesigned my website. Now I'm slowing migrating project experience and work experience over.
About Me
My name is James Guo 过仲懿. I am a Wuxiness (无锡人🇨🇳) living in 🇺🇸. My research focused on applying AI and statistical methods to solve health-related problems.
My personal life:
- laid-back body building
- laid-back swimming
- MORE on my Instagram ↗!
Please keep in touch. Thanks!
Education
Publications
[1] Shin, J.‡, Brady, E.‡, Chen, C., Lauderdale, K., Agrawal, A., Zhang, Y., Jiang, X., Nambiar, P., Herbert, J., Mallen, D., Ly, K., Guo, Z., Sant, C., Thomas, R., Miller, S., Cobos, I., Palop, J.. APOE4 and Aβ synergize to drive neuronal network dysfunction and lysosomal-ER proteostasis dysregulation in the preclinical stages of Alzheimer’s disease, Nature Neuroscience, 2025. Submitted ↗
[2] Poster session presenter and first author, Causal effect of type II diabetes on prostate cancer in the East Asian population: A two-sample Mendelian randomization study, AACR Special Conference: Aging and Cancer, 2022. Published ↗
Manuscripts in preparation
[1] (Aiming for BMC Medicine) Guo, Z.‡, Chen, D.‡, Stopsack, K. H., Soule, P., Ajit, D., Ramamoorthy, P., Hoffmann, T. J., Chan, J. M., Mucci L. A., Graff, R. E. (2025). Metabolomic Disparities Between Black and Non-Hispanic White Men with Metastatic Hormone-Sensitive Prostate Cancer: A Pilot Study.
[2] (Aiming for Cell) Qu, P., Wang, T., Jessa, S., Guo, Z., Guo, H., Purmann, C., Monte, E., Jiang, L., Yang, X., Zhou, B., Kundu, S., Kundaje, A., Wong, W., Hallmayer, J. F., Urban, A. E., Snyder, M. P.. Multi-modal functional genomics analysis of bipolar disorder and schizophrenia. (Title is tentative.)
[3] (Aiming for Nature Neuroscience) Sant, C., Guo, Z., Corces, M. R.. Preventing false discoveries in Alzheimer’s disease single-cell sequencing data using permutation testing. (Title is tentative.)
‡ indicates co-first authorship.
Experience
Research
I was very fortunate to join the PsychENCODE project at Prof. Anshul Kundaje's lab. In this project, I analyzed scRNA-seq and scATAC-seq multiome data of 3 brain regions from PsychENCODE to investigate the pathogenesis of schizophrenia (SCZ) and bipolar disorder (BPD).
Since chromatin accessibility and gene regulation are highly cell-type-specific, this project aims to analyze them at single-cell resolution. The overarching goal is to understand how variations in chromatin accessibility and gene expression across different cell types contribute to the pathogeneses of SCZ and BPD compared to controls.
Here is what I have done:
- Pre-processed the single-cell multiome dataset using quality control, dimension reduction, annotation with reference, cell clustering, doublet removal, and marker gene detection.
- Conducted differential gene expression and peak accessibility analyses (pseudobulk: DESeq2; single-cell: Wilcoxon rank-sum test and MAST) across cell types by sex and disease.
- Creatively visualized and validated differential patterns for all detected genes and peaks, presenting findings to collaborators.
Here is what I will do:
- Perform gene ontology analysis to elucidate the biological pathways associated with the identified differentially expressed genes.
- Cross-reference cell-type-specific ChromBPNet ↗ outputs to address two key questions:
- Which disease variants are causal for SCZ and BP
- How mutations impact chromatin accessibility and, consequently, gene regulation
Representative packages I have used in this project:
- R: Seurat, ArchR, Signac, Azimuth, DoubletFinder, EnsDb, SingleCellExperiment, DESeq2
- Python: CellBender, chrombpnet
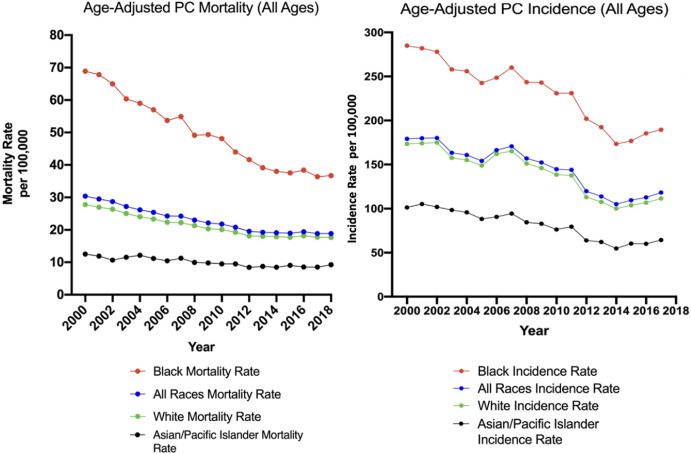
Black men experience significantly higher incidence and mortality rates of prostate cancer compared to White men. This project aims to investigate this racial disparity through LC-MS metabolomics.
The cohort included 17 Black men and 17 White men in the United States, from the IRONMAN Registry ↗.
Here is what I have done:
- Discovered key contributors through chemical similarity enrichment analysis (ChemRICH) by designing and implementing three methods: sub-pathway information, correlation modules, and predicted Medical Subject Headings (MeSH) classes.
- Found corroborative contributing metabolites using Principle Component Analysis (PCA), Partial Least Squares Discriminant Analysis (PLS-DA), Random Forest, Support Vector Machine (SVM), logistic regression, quantitative set enrichment, and pathway analysis using MetaboAnalystR.
- Identified upregulated/downregulated compounds influenced by aging through differential expression analysis using limma, after network construction and module detection using weighted gene co-expression network analysis (WGCNA), motivating research into aging's role.
The manuscript is in preparation. It will also be my thesis for my Master degree.
Representative R packages: tidyverse, ggplot2, ChemRICH, WGCNA, MetaboAnalystR, limma
Teaching
- Led discussions, graded homework and exams, held office hours, and proofread assignments and solutions before releasing.
- Graded homework and exams.
- Led discussions, graded homework and exams, held office hours, and communicated between professors and students.
- Created and stabilized knockout mutations on target gene of fruit flies using CRISPR/Cas9.
- Assisted with designing and cloning primers with sgRNA and guided 20 students in analyzing mutations vs. wildtype on the UCSC Genome Browser and in locating sgRNA transgenes.
- Directed me to computational work (sorry let's just be real)
Class Projects
to be updated...
Work

- Pioneered an R package implementing a permutation-test pipeline to reduce false positives in scRNA-seq differential gene expression analysis of Alzheimer’s disease to identify therapeutic targets, integrating eight public Synapse datasets to demonstrate real-world use.
- Applied AlphaGenome to study how genetic variants of the MAPT gene alter splicing patterns.
- Removed doublets, performed dimensionality reduction & clustering of scATAC-seq data in ArchR.
- Communicated statistical findings and effective visualizations clearly to diverse audiences.

- Revised the Enhanced Interests feature using SwiftUI, allowing users to personalize tags with text and emojis, enabling better expression of personalities.
- Improved code efficiency by replacing UIKit with SwiftUI and streamlined A/B testing.

- Extracted sales statistics from e-commerce platforms using web scraping in Python.
- Advised marketing strategies with an evidence-based report that built linear regression models using Sklearn to forecast sales trends by analyzing customer shopping patterns across multiple product categories, and included appealing visualizations created with Matplotlib in Python.
Technical Summary
R
Python
Java

Javascript
Swift
HTML
CSS
LaTeX
ipynb
RStudio
Overleaf

VS Code

Pycharm

Xcode
SAS OnDemand
UNIX

Terminal

Secure Shell

AWS EC2

Conda Env
Git

GitHub

Photoshop

Procreate

After Effect